interactive notebooks
 |  |  |
pyAgrum can easily interact with other applications. In this notebook, we propose for example some application tracks with notebook ipywidgets to make the exploration of graphical models and their inferences more interactive.
import pyagrum as gumimport pyagrum.lib.notebook as gnbListeners and progress bars
Section titled “Listeners and progress bars”import globimport os.pathfrom tqdm.auto import tqdm
class TqdmProgressBarLoadListener: def __init__(self, filename: str): self.pbar = tqdm(total=100, desc=filename, bar_format="{desc}: {percentage:3.0f}%|{bar}|")
def update(self, progress): if progress == 200: self.pbar.close() else: self.pbar.update() self.pbar.display()
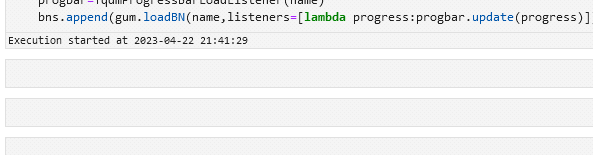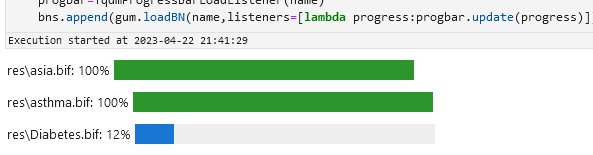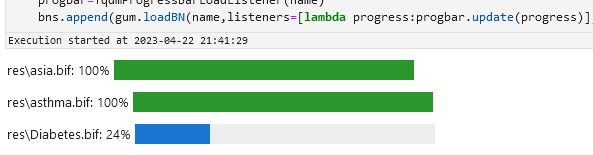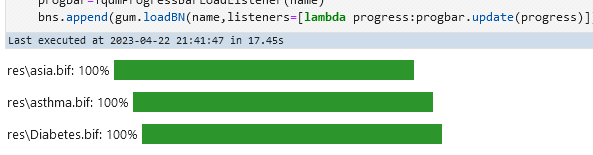
bns = {}for ext in ["dsl", "bif"]: for name in glob.glob(f"res/*.{ext}"): progbar = TqdmProgressBarLoadListener(name) bns[os.path.basename(name)] = gum.loadBN(name, listeners=[lambda progress: progbar.update(progress)])res/alarm.dsl: 0%| |
res/asia.bif: 0%| |
res/Diabetes.bif: 0%| |
res/asthma.bif: 0%| |Which should give you something like
Animated graphs
Section titled “Animated graphs”ipywidget can be used with different types of objects. Let’s say that you have a class that show the arcs of a Bayesian network only the mutual information of this arc is above a certain threshold:
import pydot as dot
class InformationViewer: def __init__(self, bn: gum.BayesNet): self.bn = bn
ie = gum.LazyPropagation(bn) self._min = float("inf") self._max = float("-inf") self._arcs = {} for x, y in bn.arcs(): nameX = bn.variable(x).name() nameY = bn.variable(y).name() ie.addJointTarget({nameX, nameY}) info = gum.InformationTheory(ie, [nameX], [nameY]) m = info.mutualInformationXY() if self._min > m: self._min = m if self._max < m: self._max = m self._arcs[x, y] = m
def min(self): return self._min
def max(self): return self._max
def showBN(self, minVal: float = 0): graph = dot.Dot(graph_type="digraph", bgcolor="transparent") bgcol = gum.config["notebook", "default_node_bgcolor"] fgcol = gum.config["notebook", "default_node_fgcolor"] for n in self.bn.names(): graph.add_node(dot.Node('"' + n + '"', style="filled", fillcolor=bgcol, fontcolor=fgcol)) for x, y in self.bn.arcs(): graph.add_edge( dot.Edge( '"' + self.bn.variable(x).name() + '"', '"' + self.bn.variable(y).name() + '"', style="invis" if self._arcs[x, y] < minVal else "", ) )
size = gum.config["notebook", "default_graph_size"] graph.set_size(size) return graph
view = InformationViewer(bns["alarm.dsl"])print(f"min={view.min()} ,max={view.max()}")gnb.sideBySide(view.showBN(0.3), view.showBN(0.5), captions=["BN filtered by $MI>0.3$", "BN filtered by $MI>0.5$"])min=7.940532588686096e-06 ,max=0.8850119269966233Now we can use this class for animation :
import ipywidgets as widgets
def interactive_view(threshold: float): return view.showBN(threshold)
widgets.interact(interactive_view, threshold=(view.min(), view.max(), (view.max() - view.min()) / 100.0));interactive(children=(FloatSlider(value=0.44250993376460596, description='threshold', max=0.8850119269966233, …Which should give you something like
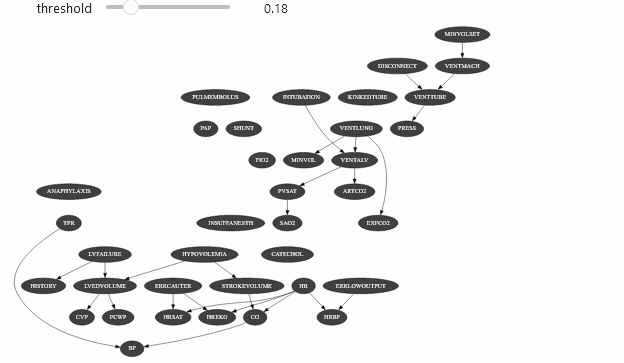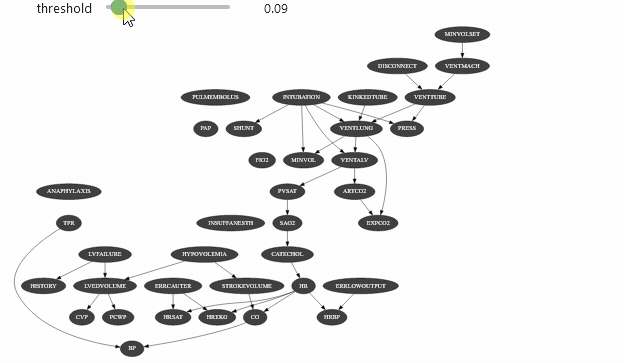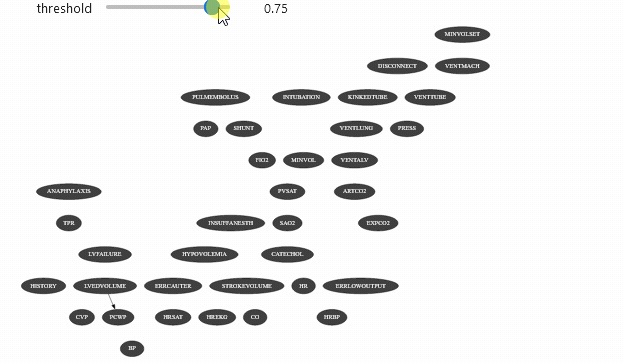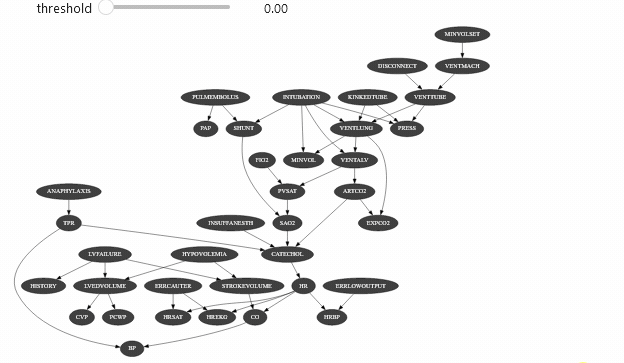
Vizualizing evidence impact
Section titled “Vizualizing evidence impact”from ipywidgets import interact, fixed
bn = bns["asia.bif"]
asia = list(bn["visit_to_Asia"].labels())smoking = list(bn["smoking"].labels())XraY = list(bn["positive_XraY"].labels())cig_ped_day = gum.RangeVariable("cigarettes_per_day", "cigarettes_per_day in [0, 10]?", 0, 10)bn.add(cig_ped_day)
@interact( bn=fixed(bn), visit_to_Asia=asia, smoking=smoking, positive_XraY=XraY, smoked_cigarettes=(cig_ped_day.minVal(), cig_ped_day.maxVal(), 1),)def evidence_impact(bn, visit_to_Asia, smoking, positive_XraY, smoked_cigarettes): evs = { "visit_to_Asia": visit_to_Asia, "smoking": smoking, "positive_XraY": positive_XraY, "cigarettes_per_day": smoked_cigarettes, } gnb.showInference(bn, evs=evs)interactive(children=(Dropdown(description='visit_to_Asia', options=('0', '1'), value='0'), Dropdown(descripti…